Anti-Ovarian Cancer Conotoxins Identified from Conus Venom
Abstract
:1. Introduction
2. Results and Discussions
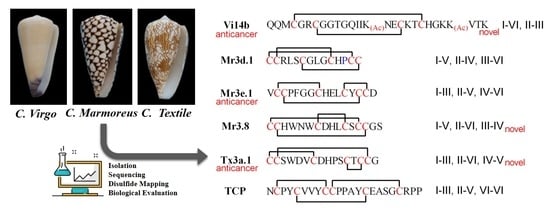
2.1. Peptide Isolation and Sequencing
2.2. Disulfide Connectivity
2.3. Blockage on Voltage-Gated Sodium Channel
2.4. Tx3a.1, Mr3.8 and Vi14b Inhibited the Proliferation of ID8 Cells
3. Materials and Methods
3.1. Venom Preparation and Peptide Isolation
3.2. Peptide Sequencing
3.3. Peptide Synthesis
3.4. Disulfide Connectivity Recognition
3.5. Patch Clamp Recording for Voltage-Gated Sodium Channel
3.6. Ovarian Cancer Cell Viability Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2019, 69, 7–34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, C.; Ralyea, C.; Lockwood, S. Ovarian Cancer: An Integrated Review. Semin. Oncol. Nurs. 2019, 35, 151–156. [Google Scholar] [CrossRef] [PubMed]
- National Cancer Institute. Surveillance, Epidemiology, and End Results Program. Cancer Stat Facts: Ovarian Cancer. Available online: https://seer.cancer.gov/statfacts/html/ovary.html (accessed on 24 August 2022).
- Jayson, G.C.; Kohn, E.C.; Kitchener, H.C.; Ledermann, J.A. Ovarian cancer. Lancet 2014, 384, 1376–1388. [Google Scholar] [CrossRef]
- Vargas, A.N. Natural history of ovarian cancer. Ecancermedicalscience 2014, 25, 465–476. [Google Scholar]
- Lheureux, S.; Braunstein, M.; Oza, A.M. Epithelial ovarian cancer: Evolution of management in the era of precision medicine. CA Cancer J. Clin. 2019, 69, 280–304. [Google Scholar] [CrossRef] [Green Version]
- Cortez, A.J.; Tudrej, P.; Kujawa, K.A.; Lisowska, K.M. Advances in ovarian cancer therapy. Cancer Chemother. Pharmacol. 2018, 81, 17–38. [Google Scholar] [CrossRef] [Green Version]
- O’Malley, D.M. New Therapies for Ovarian Cancer. J. Natl. Compr. Canc. Netw. 2019, 17, 619–621. [Google Scholar]
- Boyd, L.R.; Muggia, F.M. Carboplatin/Paclitaxel Induction in Ovarian Cancer: The Finer Points. Oncology 2018, 32, 418–420, 422–424. [Google Scholar]
- Buczek, O.; Bulaj, G.; Olivera, B.M. Conotoxins and the posttranslational modification of secreted gene products. Cell Mol. Life Sci. 2005, 62, 3067–3079. [Google Scholar] [CrossRef]
- Gao, B.; Peng, C.; Yang, J.; Yi, Y.H.; Zhang, J.Q.; Shi, Q. Cone snails: A big store of conotoxins for novel drug discovery. Toxins 2017, 9, 397. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Antunes, A. Biomedical Potential of the Neglected Molluscivorous and Vermivorous Conus Species. Mar. Drugs. 2022, 20, 105. [Google Scholar] [CrossRef]
- Robinson, S.D.; Norton, R.S. Conotoxin gene superfamilies. Mar. Drugs 2014, 12, 6058–6101. [Google Scholar] [CrossRef] [Green Version]
- Han, T.S.; Teichert, R.W.; Olivera, B.M.; Bulaj, B. Conus venoms—A rich source of peptide-based therapeutics. Curr. Pharm. Des. 2008, 14, 2462–2479. [Google Scholar] [CrossRef]
- Vetter, I.J.; Lewis, R. Therapeutic potential of cone snail venom peptides (conopeptides). Curr. Top Med. Chem. 2012, 12, 1546–1552. [Google Scholar] [CrossRef]
- Quik, M.; Wonnacott, S. α6β2* and α4β2* nicotinic acetylcholine receptors as drug targets for Parkinson’s disease. Pharmacol. Rev. 2011, 63, 938–966. [Google Scholar] [CrossRef] [Green Version]
- Becchetti, A.; Aracri, P.; Meneghini, S.; Brusco, S.; Amadeo, A. The role of nicotinic acetylcholine receptors in autosomal dominant nocturnal frontal lobe epilepsy. Front. Physiol. 2015, 6, 22–35. [Google Scholar] [CrossRef] [Green Version]
- Lewis, R.J.; Dutertre, S.; Vetter, I.; Christie, M.J. Conus venom peptide pharmacology. Pharmacol. Rev. 2012, 64, 259–298. [Google Scholar] [CrossRef]
- Gallo, A.; Boni, R.; Tosti, E. Neurobiological activity of conotoxins via sodium channel modulation. Toxicon 2020, 187, 47–56. [Google Scholar] [CrossRef]
- Dhiman, V.; Pant, D. Human health and snails. J. Immunoass. Immunochem. 2021, 42, 211–235. [Google Scholar] [CrossRef]
- Fu, Y.; Li, C.; Dong, S.; Wu, Y.; Zhangsun, D.; Luo, S. Discovery Methodology of Novel Conotoxins from Conus Species. Mar. Drugs 2018, 16, 417. [Google Scholar] [CrossRef] [Green Version]
- Pope, J.E.; Deer, T.R. Ziconotide: A clinical update and pharmacologic review. Expert Opin. Pharmacother. 2013, 14, 957–966. [Google Scholar] [CrossRef]
- Jin, A.H.; Muttenthaler, M.; Dutertre, S.; Himaya, S.W.A.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Conotoxins: Chemistry and Biology. Chem. Rev. 2019, 119, 11510–11549. [Google Scholar] [CrossRef]
- Peng, C.; Liu, L.; Shao, X.; Chi, C.; Wang, C. Identification of a novel class of conotoxins defined as V-conotoxins with a unique cysteine pattern and signal peptide sequence. Peptides 2008, 29, 985–991. [Google Scholar] [CrossRef]
- Wang, Q.; Jiang, H.; Han, Y.H.; Yuan, D.D.; Chi, C.W. Two different groups of signal sequence in M-superfamily conotoxins. Toxicon 2008, 51, 813–822. [Google Scholar] [CrossRef]
- Dutertre, S.; Jin, A.H.; Kaas, Q.; Jones, A.; Alewood, P.F.; Lewis, R.J. Deep venomics reveals the mechanism for expanded peptide diversity in cone snail venom. Mol. Cell. Proteom. 2013, 12, 312–329. [Google Scholar] [CrossRef] [Green Version]
- Tayo, L.L.; Lu, B.; Cruz, L.J.; Yates, J.R., 3rd. Proteomic analysis provides insights on venom processing in Conus textile. J. Proteome Res. 2010, 9, 2292–2301. [Google Scholar] [CrossRef] [Green Version]
- Corpuz, G.P.; Jacobsen, R.B.; Jimenez, E.C.; Watkins, M.; Walker, C.; Colledge, C.; Garrett, J.E.; McDougal, O.; Li, W.; Gray, W.R.; et al. Definition of the M-conotoxin superfamily: Characterization of novel peptides from molluscivorous Conus venoms. Biochemistry 2005, 44, 8176–8186. [Google Scholar] [CrossRef]
- Han, Y.H.; Wang, Q.; Jiang, H.; Liu, L.; Xiao, C.; Yuan, D.D.; Shao, X.X.; Dai, Q.Y.; Cheng, J.S.; Chi, C.W. Characterization of novel M-superfamily conotoxins with new disulfide linkage. FEBS J. 2006, 273, 4972–4982. [Google Scholar] [CrossRef]
- Gngora-Bentez, M.; Tulla-Puche, J.; Albericio, F. Multifaceted roles of disulfide bonds. Peptides as therapeutics. Chem. Rev. 2014, 114, 901–926. [Google Scholar] [CrossRef]
- Luo, S.; Zhangsun, D.; Harvey, P.J.; McIntosh, J.M. Cloning, synthesis, and characterization of αO-conotoxin GeXIVA, a potent α9α10 nicotinic acetylcholine receptor antagonist. Proc. Natl. Acad. Sci. USA 2015, 112, E4026–E4035. [Google Scholar] [CrossRef] [Green Version]
- Akondi, K.B.; Muttenthaler, M.; Dutertre, S.; Kaas, Q.; Craik, D.J.; Lewis, R.J.; Alewood, P.F. Discovery, synthesis, and structure–activity relationships of conotoxins. Chem. Rev. 2014, 114, 5815–5847. [Google Scholar] [CrossRef] [PubMed]
- Ueberheide, B.M.; Fenyo, D.; Alewood, P.F.; Chait, B.T. Rapid sensitive analysis of cysteine rich peptide venom components. Proc. Natl. Acad. Sci. USA 2009, 106, 6910–6915. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McDougal, O.M.; Turner, M.W.; Ormond, A.J.; Poulter, C.D. Three-dimensional structure of conotoxin tx3a: An M-1 branch peptide of the M-superfamily. Biochemistry 2008, 47, 2826–2832. [Google Scholar] [CrossRef] [PubMed]
- Dobson, R.; Collodoro, M.; Gilles, N.; Turtoi, A.; Pauw, E.D.; Quinton, L. Secretion and maturation of conotoxins in the venom ducts of Conus textile. Toxicon 2012, 60, 1370–1379. [Google Scholar] [CrossRef]
- Cruz, L.J.; Ramilo, C.A.; Corpuz, G.P.; Olivera, B.M. Conus peptides: Phylogenetic range of biological activity. Biol. Bull. 1992, 183, 159–164. [Google Scholar] [CrossRef]
- Cannon, S.C. Sodium channelopathies of skeletal muscle. Handb. Exp. Pharmacol. 2018, 246, 309–330. [Google Scholar]
- Markgraf, R.; Leipold, E.; Schirmeyer, J.; Paolini-Bertrand, M.; Hartley, O.; Heinemann, S.H. Mechanism and molecular basis for the sodium channel subtype specificity of µ-conopeptide CnIIIC. Br. J. Pharmacol. 2012, 167, 576–586. [Google Scholar] [CrossRef] [Green Version]
- Hameed, S. Nav1. 7 and Nav1. 8: Role in the pathophysiology of pain. Mol. Pain 2019, 15, 1744806919858801. [Google Scholar] [CrossRef] [Green Version]
- Ekberg, J.; Jayamanne, A.; Vaughan, C.W.; Lewis, R.J. μO-conotoxin MrVIB selectively blocks Nav1.8 sensory neuron specific sodium channels and chronic pain behavior without motor deficits. Proc. Natl. Acad. Sci. USA 2006, 103, 17030–17035. [Google Scholar] [CrossRef] [Green Version]
- Breipohl, G.; Knolle, J.; Stuber, W. Synthesis and application of acid labile anchor groups for the synthesis of peptide amides by Fmoc-solid-phase peptide synthesis. Int. J. Pept. Protein Res. 1989, 34, 262–267. [Google Scholar] [CrossRef]
- Heimer, P.; Tietze, A.A.; Bäuml, C.A.; Resemann, A.; Mayer, F.J.; Suckau, D.; Ohlenschläger, O.; Tietze, D.; Imhof, D. Conformational μ-conotoxin PIIIA isomers revisited: Impact of cysteine pairing on disulfide-bond assignment and structure elucidation. Anal. Chem. 2018, 90, 3321–3327. [Google Scholar] [CrossRef]
- Weiser, T. Comparison of the effects of four Na+ channel analgesics on TTX-resistant Na+ currents in rat sensory neurons and recombinant Nav1.2 channels. Neurosci. Lett. 2006, 395, 179–184. [Google Scholar] [CrossRef]
- Mazurek, N.; Byrd, J.C.; Sun, Y.; Ueno, S.; Bresalier, R.S. A galectin-3 sequence polymorphism confers TRAIL sensitivity to human breast cancer cells. Cancer 2011, 117, 4375–4380. [Google Scholar] [CrossRef]









| Name | Sequence | Origin | Disulfide Bridge | Reference |
|---|---|---|---|---|
| Vi14b | QQMCGRCGGTGQIIK(Ac)NECKTCHGKK(Ac) VTK | C. virgo | 2 pairs | This work |
| Vi002 | LSSGATALSGVPRLTKPAGRLTTTTVAVAF * | C. virgo | none | This work |
| Vi003 | NTESTKGESLLGK * | C. virgo | none | This work |
| Vi15a | DCTTCAGEECCGRCTCPWGDNCSCTEW-(nh2) | C. virgo | 4 pairs | [24] |
| Mr3d.1 | CCRLSCGLGCHPCC | C. marmoreus | 3 pairs | This work |
| Mr3e.1 | VCCPFGGCHELCYCCD | C. marmoreus | 3 pairs | This work |
| Mr3.8 | CCHWNWCDHLCSCCGS | C. marmoreus | 3 pairs | [25,26] |
| Tx3a.1 | CCSWDVCDHPSCTCCG | C. texile | 3 pairs | This work |
| TCP | NCPYCVVYCCPPAYCEASGCRPP | C. texile | 3 pairs | [27] |
| Sequence | ya/yb | Sequence | ya/yb | Sequence | ya/yb |
|---|---|---|---|---|---|
| RCGGT | 475.2082 | TGQIIK(Ac) | 683.4087 | GKK(Ac) | 356.2292 |
| RCGGTG | 504.2347 | IIK(Ac) | 397.2809 | GKK(Ac)V | 427.3027 |
| CGGTGQ | 504.1871 | IK(Ac)N | 398.2398 | GKK(Ac)VT | 528.3504 |
| CGGTGQI | 617.2712 | IK(Ac)NE | 527.2824 | KK(Ac)V | 398.2762 |
| TGQI | 372.2241 | HGK | 323.1826 | K(Ac)VT | 371.2289 |
| IK(Ac) | 284.1969 | GK | 186.1237 |
| Sequence | ya/yb | Sequence | ya/yb | Sequence | ya/yb |
|---|---|---|---|---|---|
| SSG | 232.0928 | GVP | 226.1550 | LTT | 316.1867 |
| SSGA | 303.1299 | PR | 226.1662 | AGRLTTT | 673.3992 |
| SSGAT | 404.1776 | LT | 187.1441 | RLTTTT | 674.3832 |
| GATA | 301.1506 | LTK | 315.2391 | TTTV | 403.2187 |
| GATAL | 386.2398 | TKPAG | 455.2613 | TTTVA | 474.2558 |
| GATALS | 473.2718 | PAG | 226.1186 | TTTTVAV | 545.3293 |
| GATALSG | 558.2882 | AGR | 285.1670 | TTVAV | 472.2766 |
| SGVP | 341.1819 | GRL | 299.2190 | VAVA | 341.2183 |
| GV | 129.1022 | LT | 187.1441 |
| Name | TP1 | TP2 |
|---|---|---|
| Mr3e.1 | 3.79 ± 0.61% on Nav1.4 | 21.89 ± 3.06% on Nav1.4 |
| Mr3.8 | 4.62 ± 1.06% on Nav1.4 | 23.61 ± 2.70% on Nav1.4 |
| Tx3a.1 | 3.17 ± 0.63% on Nav1.4 | 24.32 ± 2.29% on Nav1.4 |
| TCP | 5.45 ± 0.39% on Nav1.8 | 21.51 ± 1.00% on Nav1.8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ju, S.; Zhang, Y.; Guo, X.; Yan, Q.; Liu, S.; Ma, B.; Zhang, M.; Bao, J.; Luo, S.; Fu, Y. Anti-Ovarian Cancer Conotoxins Identified from Conus Venom. Molecules 2022, 27, 6609. https://doi.org/10.3390/molecules27196609
Ju S, Zhang Y, Guo X, Yan Q, Liu S, Ma B, Zhang M, Bao J, Luo S, Fu Y. Anti-Ovarian Cancer Conotoxins Identified from Conus Venom. Molecules. 2022; 27(19):6609. https://doi.org/10.3390/molecules27196609
Chicago/Turabian StyleJu, Shuang, Yu Zhang, Xijun Guo, Qinghui Yan, Siyi Liu, Bokai Ma, Mei Zhang, Jiaolin Bao, Sulan Luo, and Ying Fu. 2022. "Anti-Ovarian Cancer Conotoxins Identified from Conus Venom" Molecules 27, no. 19: 6609. https://doi.org/10.3390/molecules27196609